Note
Click here to download the full example code or to run this example in your browser via Binder
Partial Dependence Plots with categorical values¶
Sigurd Carlsen Feb 2019 Holger Nahrstaedt 2020
Plot objective now supports optional use of partial dependence as well as different methods of defining parameter values for dependency plots.
print(__doc__)
import sys
from skopt.plots import plot_objective
from skopt import forest_minimize
import numpy as np
np.random.seed(123)
import matplotlib.pyplot as plt
import numpy as np
from sklearn.datasets import load_breast_cancer
from sklearn.tree import DecisionTreeClassifier
from sklearn.model_selection import cross_val_score
from skopt.space import Integer, Categorical
from skopt import plots, gp_minimize
from skopt.plots import plot_objective
objective function¶
Here we define a function that we evaluate.
def objective(params):
clf = DecisionTreeClassifier(
**{dim.name: val for dim, val in
zip(SPACE, params) if dim.name != 'dummy'})
return -np.mean(cross_val_score(clf, *load_breast_cancer(return_X_y=True)))
Bayesian optimization¶
SPACE = [
Integer(1, 20, name='max_depth'),
Integer(2, 100, name='min_samples_split'),
Integer(5, 30, name='min_samples_leaf'),
Integer(1, 30, name='max_features'),
Categorical(list('abc'), name='dummy'),
Categorical(['gini', 'entropy'], name='criterion'),
Categorical(list('def'), name='dummy'),
]
result = gp_minimize(objective, SPACE, n_calls=20)
Partial dependence plot¶
Here we see an example of using partial dependence. Even when setting n_points all the way down to 10 from the default of 40, this method is still very slow. This is because partial dependence calculates 250 extra predictions for each point on the plots.
_ = plot_objective(result, n_points=10)
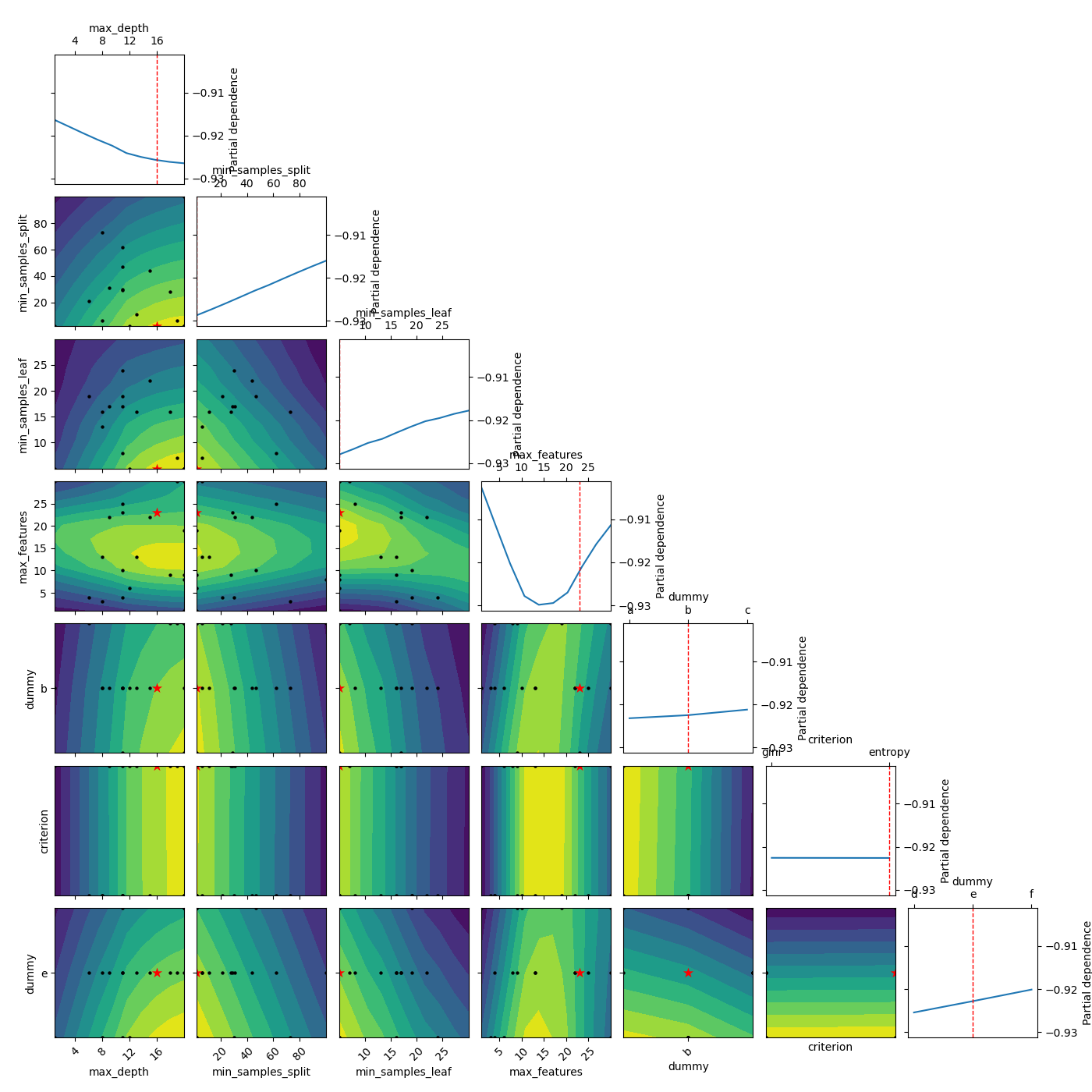
Plot without partial dependence¶
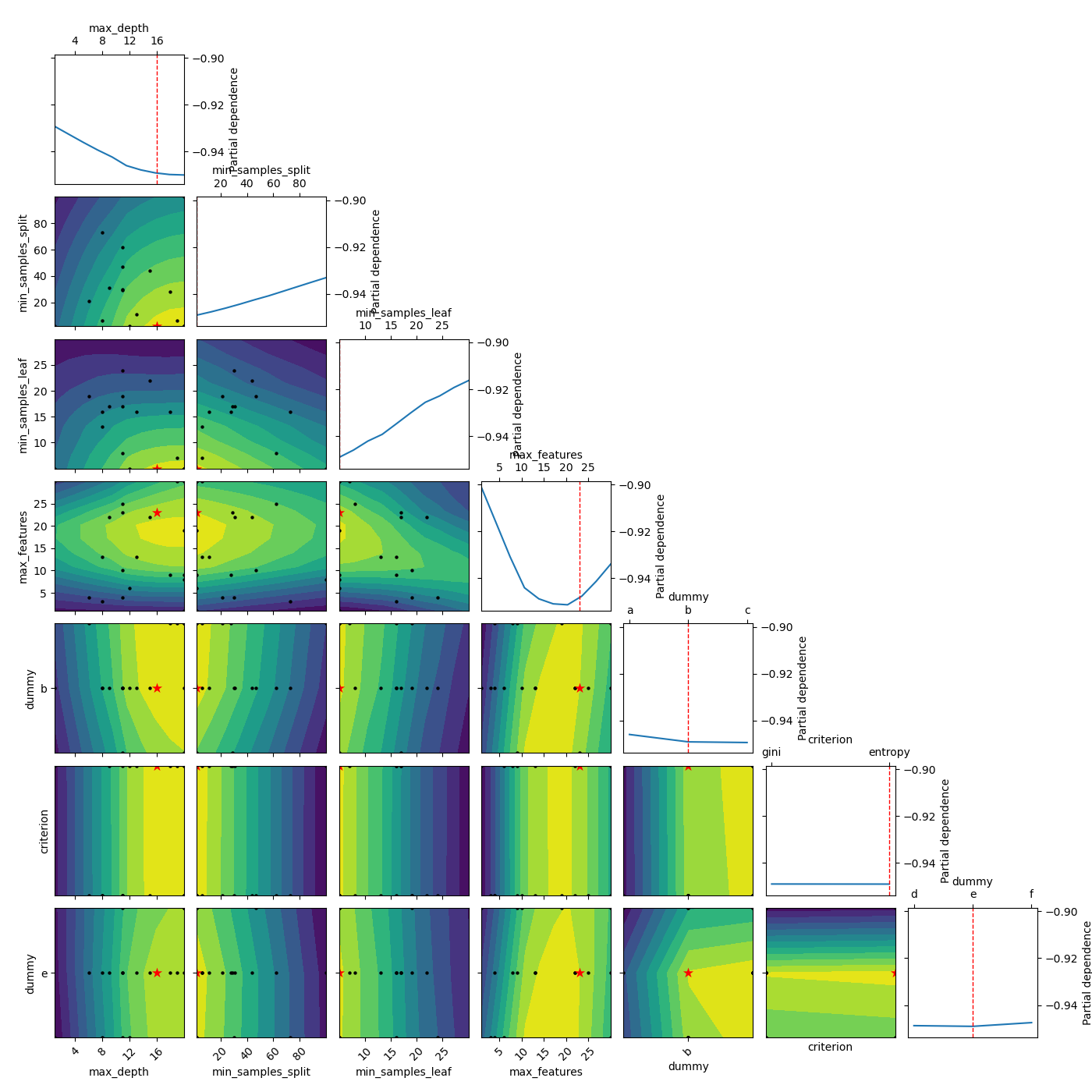
Here we plot without partial dependence. We see that it is a lot faster. Also the values for the other parameters are set to the default “result” which is the parameter set of the best observed value so far. In the case of funny_func this is close to 0 for all parameters.
_ = plot_objective(result, sample_source='result', n_points=10)
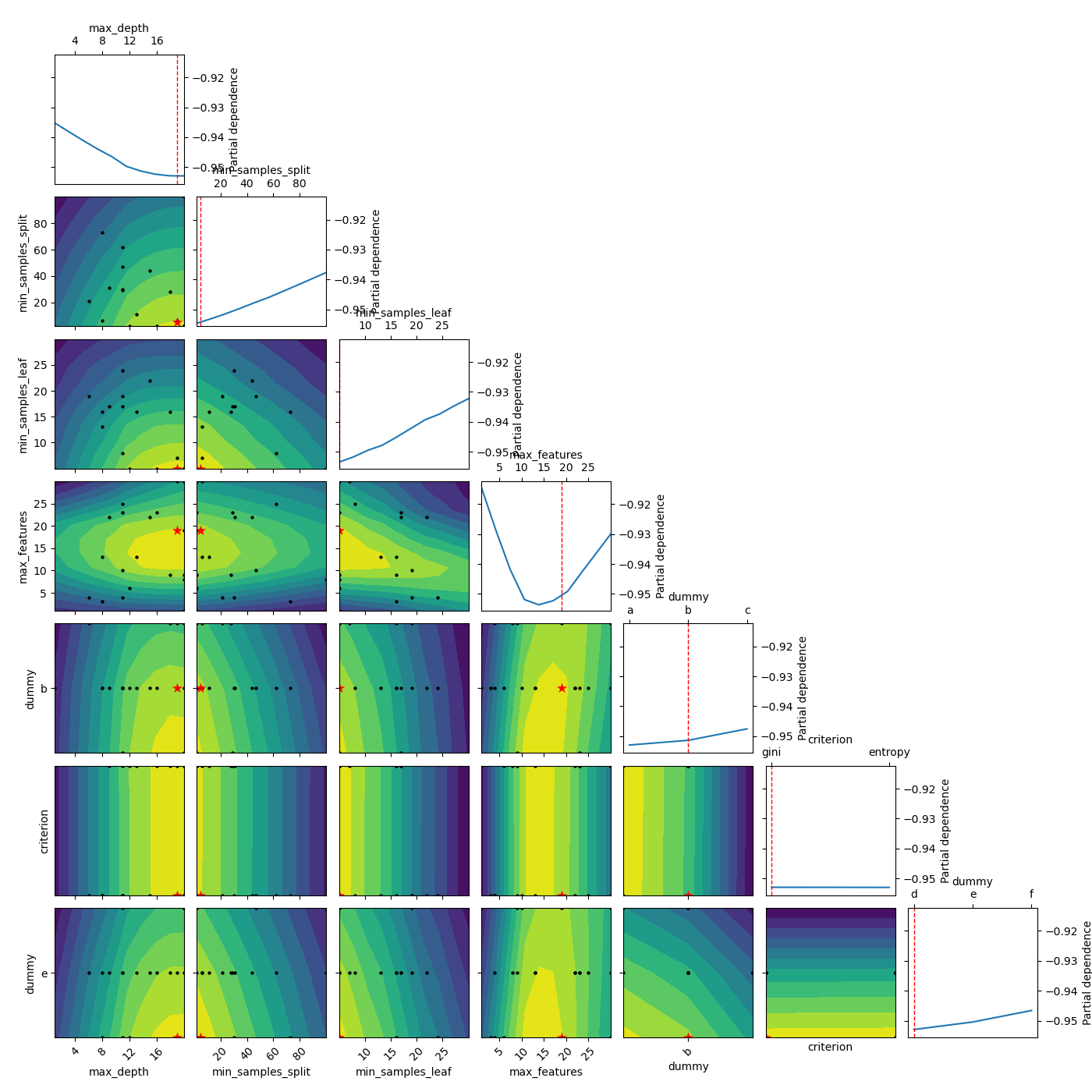
Modify the shown minimum¶
Here we try with setting the other parameters to something other than
“result”. When dealing with categorical dimensions we can’t use
‘expected_minimum’. Therefore we try with “expected_minimum_random”
which is a naive way of finding the minimum of the surrogate by only
using random sampling. n_minimum_search sets the number of random samples,
which is used to find the minimum
_ = plot_objective(result, n_points=10, sample_source='expected_minimum_random',
minimum='expected_minimum_random', n_minimum_search=10000)
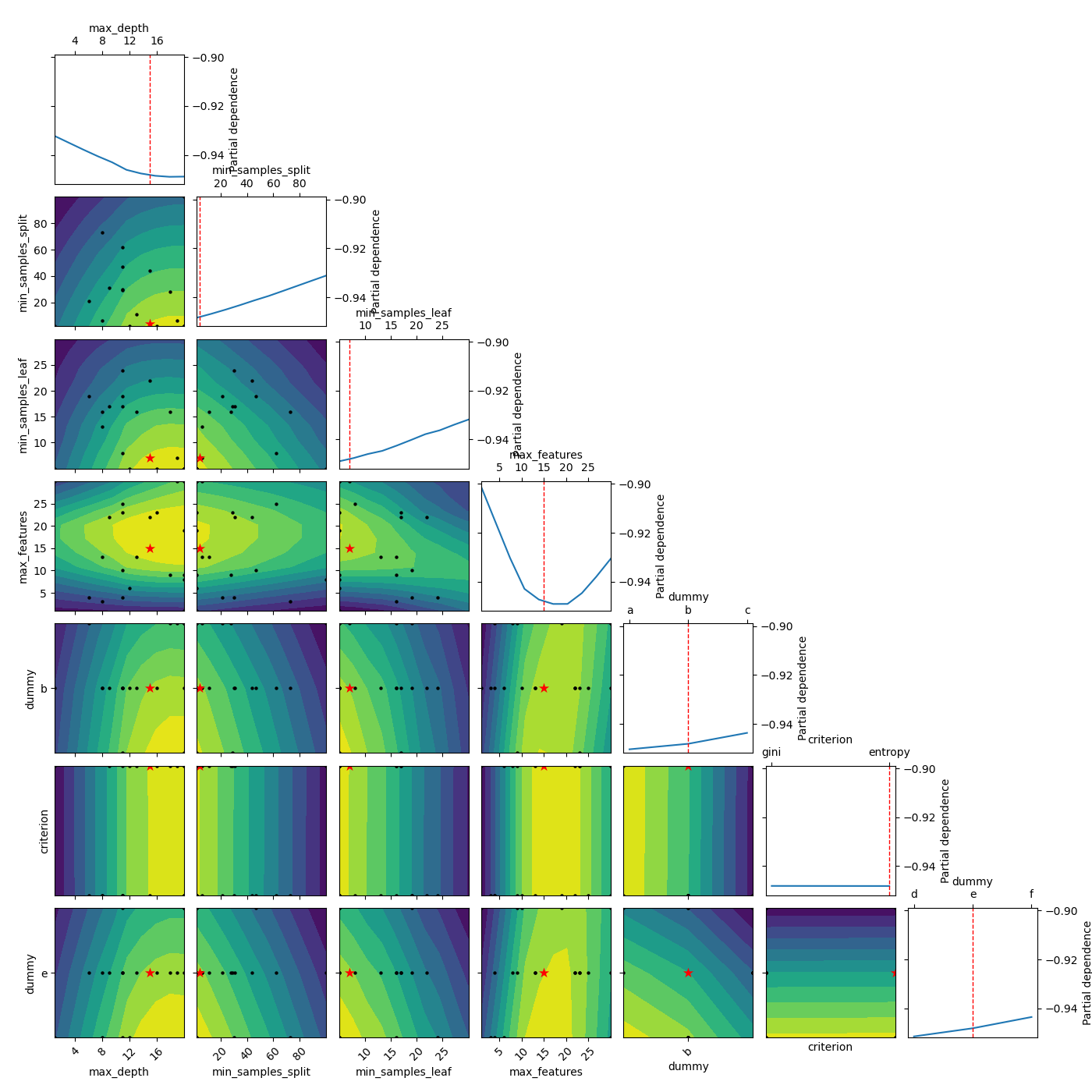
Set a minimum location¶
Lastly we can also define these parameters ourselfs by parsing a list as the pars argument:
_ = plot_objective(result, n_points=10, sample_source=[15, 4, 7, 15, 'b', 'entropy', 'e'],
minimum=[15, 4, 7, 15, 'b', 'entropy', 'e'])
Total running time of the script: ( 0 minutes 13.991 seconds)
Estimated memory usage: 52 MB
